Research
Imaging breakthrough leads to further understanding of genome structure and function
January 06, 2021
The imaging platform combines four techniques to provide imaging and sensing capabilities.
A combination of novel techniques helped shed light on how the genome is constructed
The way cells, organisms, and human bodies perform is dictated by much more than genetics. While each cell in a body has the exact same set of genes, the cells themselves can be very different. What we do and how our organs function is dependent not just on our cells but how the genes within them are expressed.
Using a combination of breakthrough imaging techniques, new research from Northwestern Engineering sheds light on how cells – including cancerous ones – are constructed at the genomic level and how that structure affects their functionality and ability to adapt to external stressors.

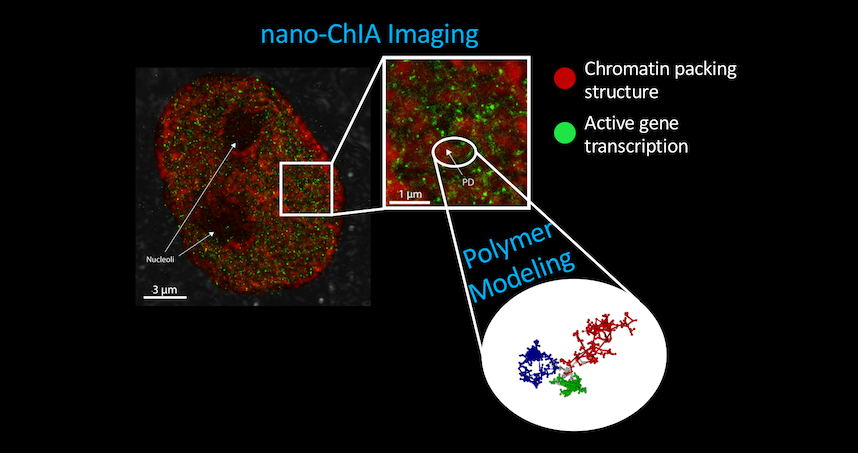
The paper “Nanoscale Chromatin Imaging and Analysis (Nano-ChIA) Platform Bridges 4D Chromatin Organization with Molecular Function” was published January 1 in the journal Science Advances.
The platform combines four techniques to provide imaging and sensing capabilities: optical super-resolution microscopy, electron tomography, and label-free, sub-diffractional optical nano-sensing. The combination allows researchers to investigate genome structure across multiple length scales in real time.
“There are a lot of interesting questions we’re able to address,” said Vadim Backman, the Walter Dill Scott Professor of Biomedical Engineering at the McCormick School of Engineering and an affiliated faculty member of the International Institute for Nanotechnology (IIN), who led the research.
Chromatin is the three-dimensional organization of the genome, and is packed into the nucleus of cells with incredible density. Cellular processes are in part regulated by the mechanisms of chromatin packing and its relationship with gene expression. In particular, chromatin packing determines how adaptable cells are, how they respond to various stresses and stimuli, and how they change while maintaining their overall “identity.”
“Having a gene does not necessarily confer a behavior,” Backman said. “At least 50 percent of what cells do is regulated outside of what the genes are.”
This is one of the most critical characteristics of life. Cells in our bodies don’t always do a single job and express the same genes. Instead, the cells must sense what is occurring in their environment and respond. Without this adaptive ability, all life forms would have quickly vanished long ago.

Armed with the capabilities provided by the new techniques, the ability to study the relationship between chromatin structure and cellular function has implications for understanding disease, including cancer. Cancer cells have a tremendous ability to evolve and change, but they are not supernatural. They continuously face challenges: they outgrow their blood supply, get attacked by the host’s immune system, and have to find ways to adapt and survive in new surroundings as they metastasize and spread through the body.
The cancer cells that survive this adaptive process – those with the most plasticity-fostering chromatin structure – are the strongest and best equipped to adjust and evade treatments. Previous research established that chromatin packing is key to their survival.
The current work shows that cellular nuclei are not randomly packed with chromatin, but rather contain sub-micron-sized domains. The internal structure of these domains regulates how genes are expressed, an insight made possible by the development of the nano-ChIA platform which allows for in-depth characterization of chromatin structure and how it relates with molecular function at all relevant length and time scales.
“Using these different platforms and combining them with a physics-based understanding of how the genome is organized allows us to capture a lot of important information,” said Ranya Virk, a PhD candidate in biomedical engineering at Northwestern and the study’s first author.
Genome structure has to be probed at the nanoscale, which is a challenging endeavor as many features cannot be imaged using conventional techniques. The structures are too big for crystallography, but too small for optical microscopy. No single technique can span all the length scales needed to look at the overall size of the chromatin chain, the sub-micron-sized domains, and even smaller units that are beneath the wavelength of light itself.
Backman was the study’s corresponding author, and the work is part of joint research between the Center for Physical Genomics and Engineering (CPGE), which Backman directs, and the Metrology in Biology initiative by Vinayak P. Dravid, Abraham Harris Professor of Materials Science and Engineering at the McCormick School of Engineering and the IIN’s Director of Global Initiatives.
“This research is an example of convergent science, is an example of why the interdisciplinary research environment at CPGE is so critical to enable complex projects like these,” Backman said.
“We are pioneering the use of scanning transmission electron microscopy (STEM) in biology and its correlation with complementary light-optical microscopy and modeling/simulation,” Dravid said. “Imagine how a brand-new telescope may open the view of the heavens. ChromSTEM may indeed prove to be that elusive imaging modality to provide further insights in the most basic and fundamental questions about what and who we are and how all this may have started.”
Yue Li, a recent graduate in both Backman’s center and Dravid’s VPD Group, was a senior leader on the project.